Publications
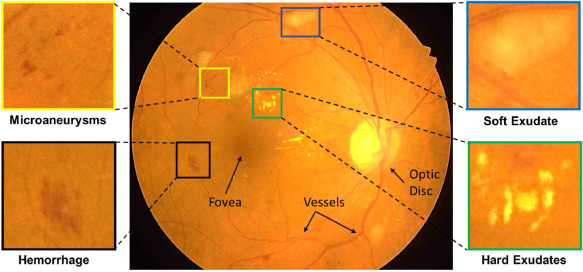
Diabetic Retinopathy (DR) is the most common cause of avoidable vision loss, predominantly affecting the working-age population across the globe. Screening for DR, coupled with timely consultation and treatment, is a globally trusted policy to avoid vision loss. However, implementation of DR screening programs is challenging due to the scarcity of medical professionals able to screen a growing global diabetic population at risk for DR. Computer-aided disease diagnosis in retinal image analysis could provide a sustainable approach for such large-scale screening effort. The recent scientific advances in computing capacity and machine learning approaches provide an avenue for biomedical scientists to reach this goal. Aiming to advance the state-of-the-art in automatic DR diagnosis, a grand challenge on “Diabetic Retinopathy – Segmentation and Grading” was organized in conjunction with the IEEE …
Prasanna Porwal, Samiksha Pachade, Manesh Kokare, Debdoot Sheet, Oindrila Saha, Rachana Sathish et al.
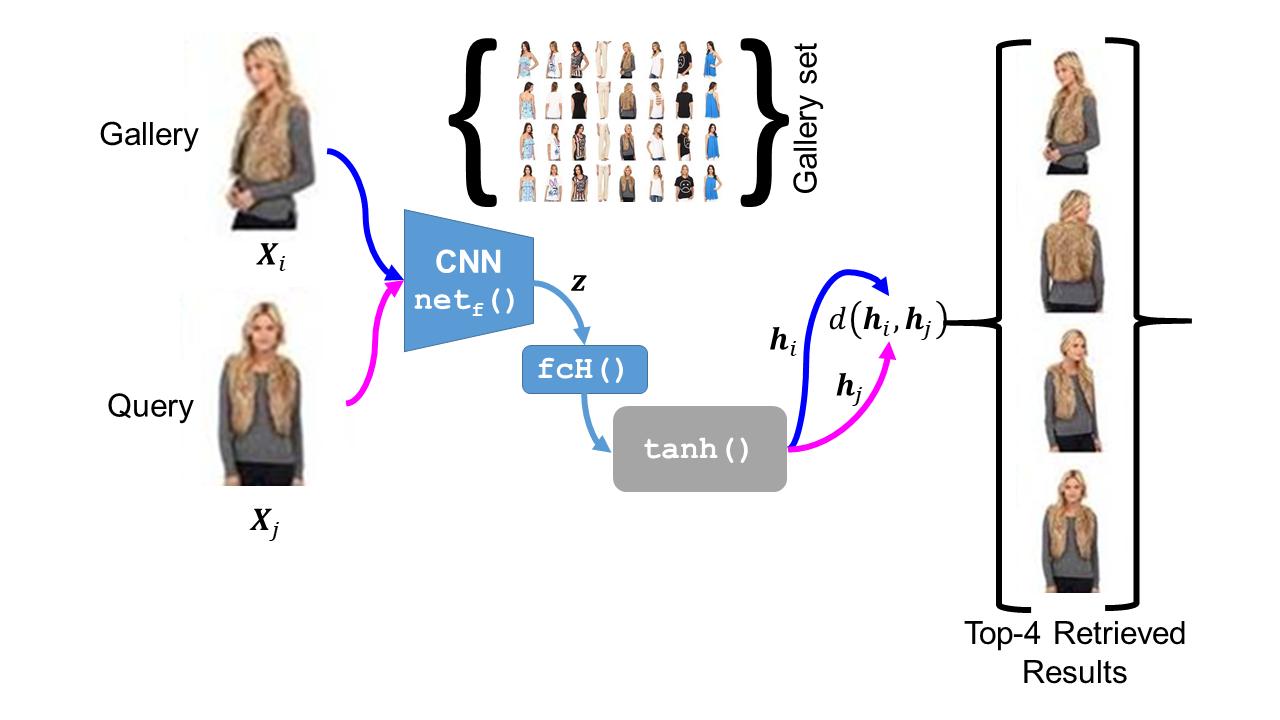
The simple approach of retrieving a closest match of a query image from one in the gallery, compares an image pair using sum of absolute difference in pixel or feature space. The process is computationally expensive, ill-posed to illumination, background composition, pose variation, as well as inefficient to be deployed on gallery sets with more than 1000 elements. Hashing is a faster alternative which involves representing images in reduced dimensional simple feature spaces. Encoding images into binary hash codes enables similarity comparison in an image-pair using the Hamming distance measure. The challenge, however, lies in encoding the images using a semantic hashing scheme that lets subjective neighbors lie within the tolerable Hamming radius. This work presents a solution employing adversarial learning of a deep neural semantic hashing network for fashion inventory retrieval. It consists of a feature extracting convolutional neural network (CNN) learned to (i) minimize error in classifying type of clothing,(ii) minimize hamming distance between semantic neighbors and maximize distance between semantically dissimilar images,(iii) maximally scramble a discriminator’s ability to identify the corresponding hash code-image pair when processing a semantically similar query-gallery image pair. Experimental validation for fashion inventory search yields a mean average precision (mAP) of 90.65% in finding the closest match as compared to 53.26% obtained by the prior art of deep Cauchy hashing for hamming space retrieval.
Saket Singh, Debdoot Sheet, Mithun Dasgupta
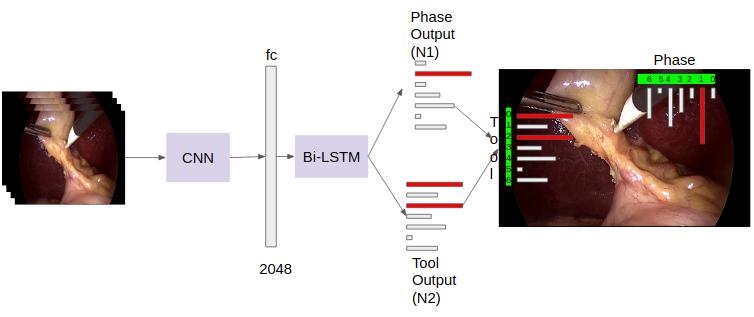
Surgical workflow analysis is of importance for understanding onset and persistence of surgical phases and individual tool usage across surgery and in each phase. It is beneficial for clinical quality control and to hospital administrators for understanding surgery planning. Video acquired during surgery typically can be leveraged for this task. Currently, a combination of convolutional neural network (CNN) and recurrent neural networks (RNN) are popularly used for video analysis in general, not only being restricted to surgical videos. In this paper, we propose a multi-task learning framework using CNN followed by a bi-directional long short term memory (Bi-LSTM) to learn to encapsulate both forward and backward temporal dependencies. Further, the joint distribution indicating set of tools associated with a phase is used as an additional loss during learning to correct for their co-occurrence in any predictions. Experimental evaluation is performed using the Cholec80 dataset. We report a mean average precision (mAP) score of 0.99 and 0.86 for tool and phase identification respectively which are higher compared to prior-art in the fie
Shanka Subhra Mondal, Rachana Sathish, Debdoot Sheet
MedImage Workshop, 2018 Indian Conference on Vision, Graphics and Image Processing
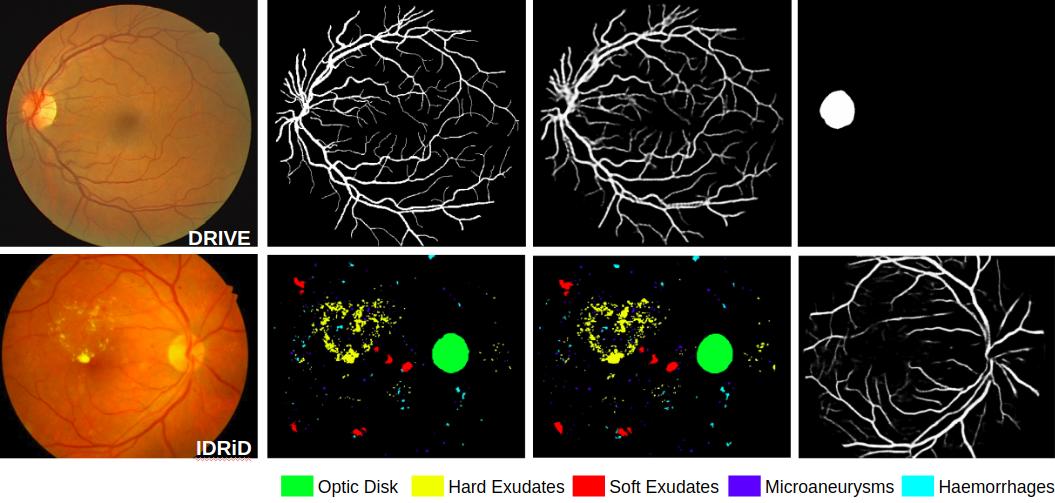
A prime challenge in building data driven inference models is the unavailability of statistically significant amount of labelled data. Since datasets are typically designed for a specific purpose, and accordingly are weakly labelled for only a single class instead of being exhaustively annotated. Despite there being multiple datasets which cumulatively represents a large corpus, their weak labelling poses challenge for direct use. As in case of retinal images which have inspired development of data driven learning based algorithms for segmenting anatomical landmarks like vessels and optic disc as well as pathologies like microaneurysms, hemorrhages, hard exudates and soft exudates; aspired to segment all using only a single fully convolutional neural network (FCN), there is no single dataset with all classes annotated. We solve this problem by training a single network using separate weakly labelled datasets. Essentially we use an adversarial learning approach over a semantic segmentation FCN, where the objectives of discriminators are to learn to (a) predict which of the classes are actually present in the input fundus image, and (b) distinguish between manual annotations vs. segmented results for each of the classes. The first discriminator works to enforce the network to segment those classes which are present in the fundus image although may not have been annotated ie all retinal images have vessels while pathology datasets may not have annotated them in the dataset. The second discriminator contributes to making the segmentation result as realistic as possible. We experimentally demonstrate using weakly labelled datasets of DRIVE …
Oindrila Saha, Rachana Sathish, Debdoot Sheet
International Conference on Medical Imaging with Deep Learning
Radiologists use various imaging modalities to aid in different tasks like diagnosis of disease, lesion visualization, surgical planning and prognostic evaluation. Most of these tasks rely on the the accurate delineation of the anatomical morphology of the organ, lesion or tumor. Deep learning frameworks can be designed to facilitate automated delineation of the region of interest in such cases with high accuracy. Performance of such automated frameworks for medical image segmentation can be improved with efficient integration of information from multiple modalities aided by suitable learning strategies. In this direction, we show the effectiveness of residual network trained adversarially in addition to a boundary weighted loss. The proposed methodology is experimentally verified on the SPES-ISLES 2015 dataset for ischaemic stroke segmentation with an average Dice coefficient of 0.881 for penumbra and 0.877 for core. It was observed that addition of residual connections and boundary weighted loss improved the performance significantly.
Ronnie Rajan, Rachana Sathish, Debdoot Sheet
2019 International Conference on Medical Imaging with Deep Learning
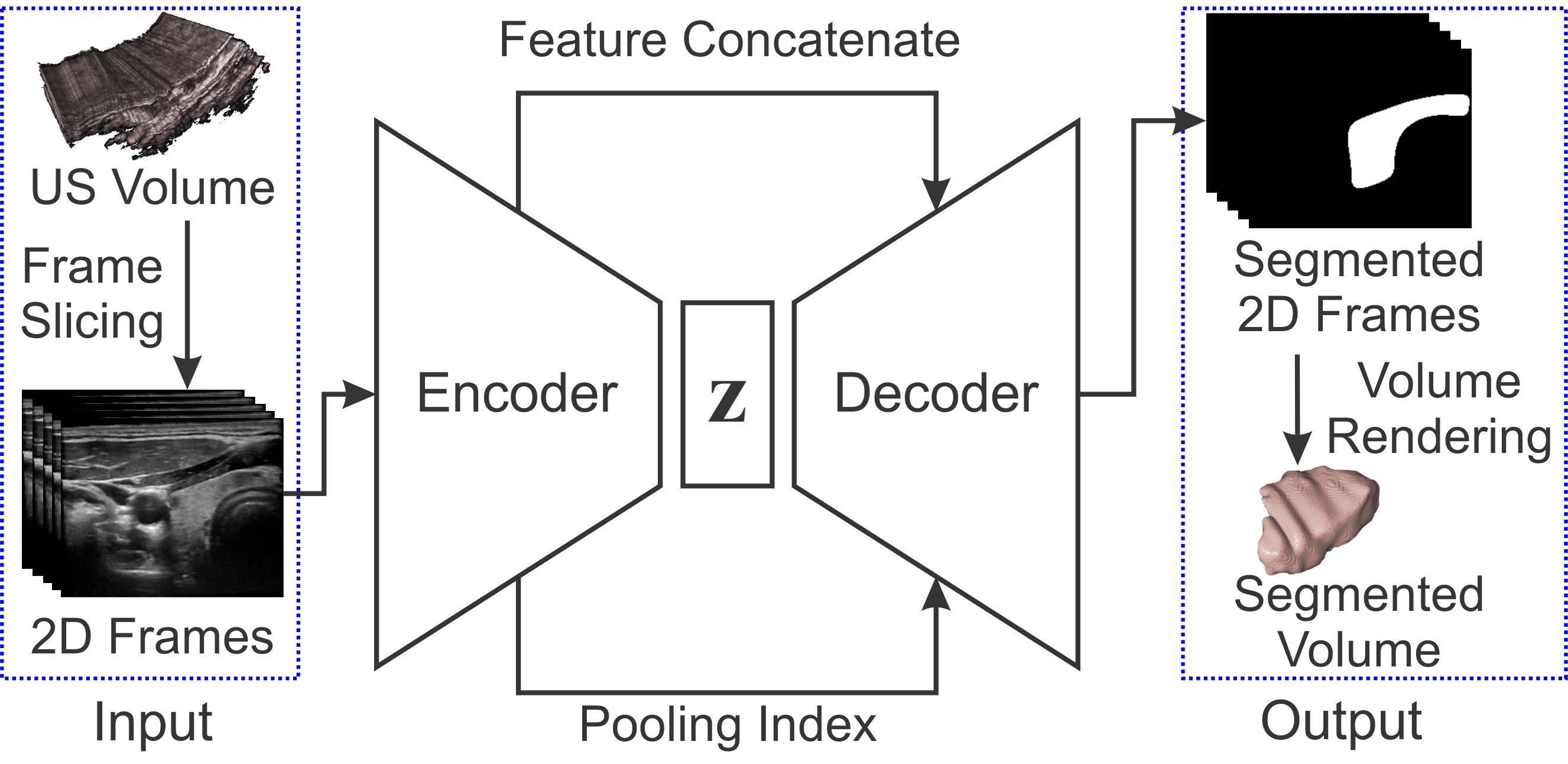
Ultrasound imaging is generally employed for real-time investigation of internal anatomy of the human body for disease identification. Delineation of the anatomical boundary of organs and pathological lesions is quite challenging due to the stochastic nature of speckle intensity in the images, which also introduces visual fatigue for the observer. This paper introduces a fully convolutional neural network based method to segment organ and pathologies in ultrasound volume by learning the spatial-relationship between closely related classes in the presence of stochastically varying speckle intensity. We propose a convolutional encoder-decoder like framework with (i) feature concatenation across matched layers in encoder and decoder and (ii) index passing based unpooling at the decoder for semantic segmentation of ultrasound volumes. We have experimentally evaluated the performance on publicly available datasets consisting of 10 intravascular ultrasound pullback acquired at 20MHz and 16freehand thyroid ultrasound volumes acquired 11-16 MHz. We have obtained a dice score of0.93±0.08 and 0.92±0.06 respectively, following a 10 -fold cross-validation experiment while processing frame of 256x384 pixel in 0.035 s and a volume of 256x384x384 voxel in 13.44 s.
Sumanth Nandamuri, Debarghya China, Pabitra Mitra, Debdoot Sheet
2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)
Adversarially trained deep neural networks have significantly improved performance of single image super resolution, by hallucinating photorealistic local textures, thereby greatly reducing the perception difference between a real high resolution image and its super resolved (SR) counterpart. However, application to medical imaging requires preservation of diagnostically relevant features while refraining from introducing any diagnostically confusing artifacts. We propose using a deep convolutional super resolution network (SRNet) trained for (i) minimising reconstruction loss between the real and SR images, and (ii) maximally confusing learned relativistic visual Turing test (rVTT) networks to discriminate between (a) pair of real and SR images (T1) and (b) pair of patches in real and SR selected from region of interest (T2). The adversarial loss of T1 and T2 while backpropagated through SRNet helps it learn to reconstruct pathorealism in the regions of interest such as white blood cells (WBC) in peripheral blood smears or epithelial cells in histopathology of cancerous biopsy tissues, which are experimentally demonstrated here. Experiments performed for measuring signal distortion loss using peak signal to noise ratio (pSNR) and structural similarity (SSIM) with variation of SR scale factors, impact of rVTT adversarial losses, and impact on reporting using SR on a commercially available artificial intelligence (AI) digital pathology system substantiate our claims.
Francis Tom, Himanshu Sharma, Dheeraj Mundhra, Tathagato Rai Dastidar, Debdoot Sheet
2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)
Ultrasound image compression by preserving speckle-based key information is a challenging task. In this paper, we introduce an ultrasound image compression framework with the ability to retain realism of speckle appearance despite achieving very high-density compression factors. The compressor employs a tissue segmentation method, transmitting segments along with transducer frequency, number of samples and image size as essential information required for decompression. The decompressor is based on a convolutional network trained to generate patho-realistic ultrasound images which convey essential information pertinent to tissue pathology visible in the images. We demonstrate generalizability of the building blocks using two variants to build the compressor. We have evaluated the quality of decompressed images using distortion losses as well as perception loss and compared it with other off the shelf solutions. The proposed method achieves a compression ratio of725:1 while preserving the statistical distribution of speckles. This enables image segmentation on decompressed images to achieve dice score of 0.89±0.11 , which evidently is not so accurately achievable when images are compressed with current standards like JPEG, JPEG 2000, WebP and BPG. We envision this frame work to serve as a roadmap for speckle image compression standards.
Debarghya China, Francis Tom, Sumanth Nandamuri, Aupendu Kar, Mukundhan Srinivasan, Pabitra Mitra, Debdoot Sheet
2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)
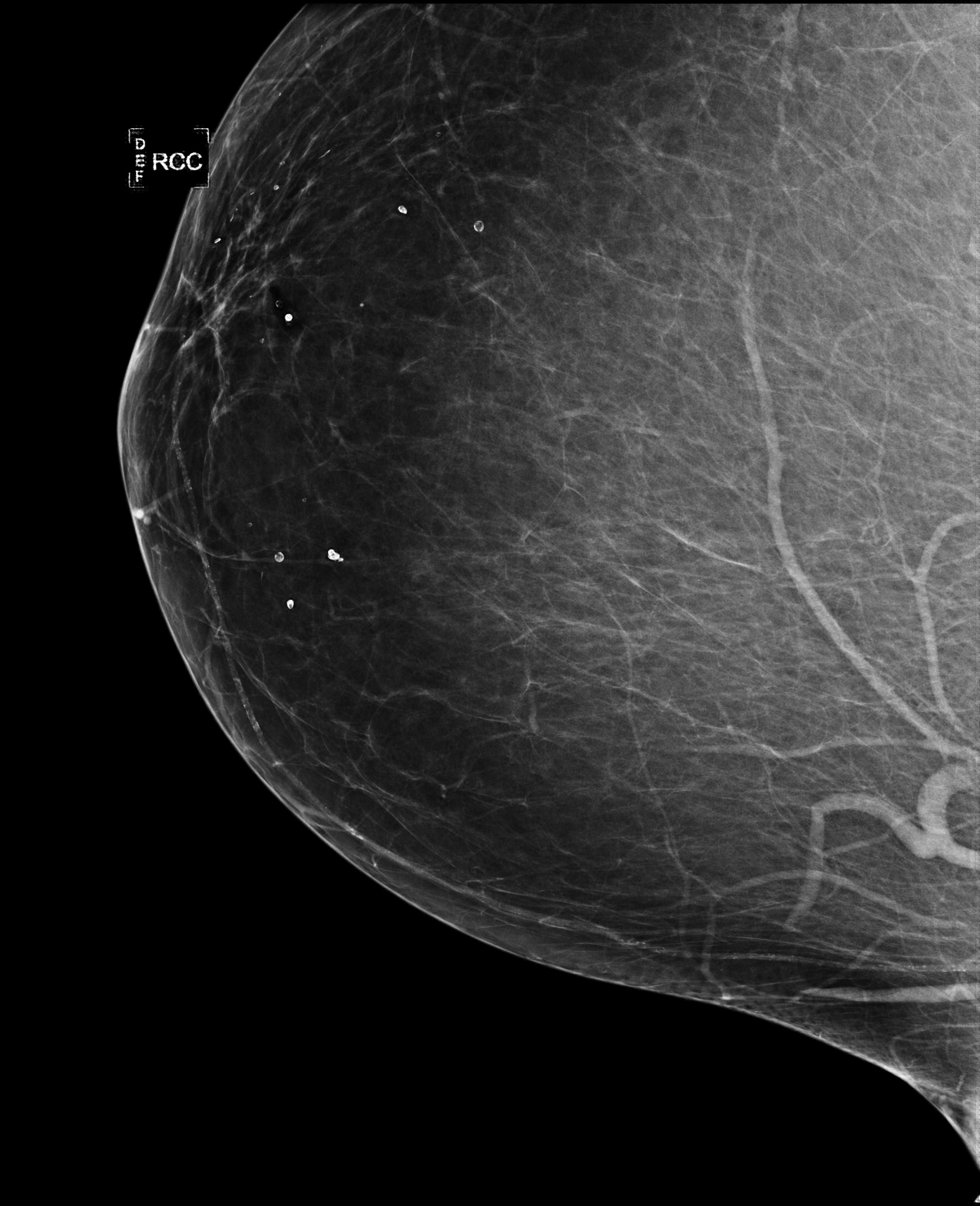
Early works on medical image compression date to the 1980’s with the impetus on deployment of teleradiology systems for high-resolution digital X-ray detectors. Commercially deployed systems during the period could compress 4,096× 4,096 sized images at 12 bpp to 2 bpp using lossless arithmetic coding, and over the years JPEG and JPEG2000 were imbibed reaching upto 0.1 bpp. Inspired by the reprise of deep learning based compression for natural images over the last two years, we propose a fully convolutional autoencoder for diagnostically relevant feature preserving lossy compression. This is followed by leveraging arithmetic coding for encapsulating high redundancy of features for further high-density code packing leading to variable bit length. We demonstrate performance on two different publicly available digital mammography datasets using peak signal-to-noise ratio (pSNR), structural similarity (SSIM) index and domain adaptability tests between datasets. At high density compression factors of> 300×(0.04 bpp), our approach rivals JPEG and JPEG2000 as evaluated through a Radiologist’s visual Turing test.
Aupendu Kar, Sri Phani Krishna Karri, Nirmalya Ghosh, Ramanathan Sethuraman, Debdoot Sheet
The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops
Full list of publications
(You may also visit the Google Scholar page)
- 2020:
-
Prasanna Porwal, Samiksha Pachade, Manesh Kokare, Debdoot Sheet, Oindrila Saha, Rachana Sathish et al., "IDRiD: Diabetic Retinopathy–Segmentation and Grading Challenge", Medical image analysis pp. 101561
-
MK Sharma, Debdoot Sheet, Prabir Kumar Biswas, "Image Embedding for Detecting Irregularity", Proceedings of 3rd International Conference on Computer Vision and Image Processing pp. 243-255
-
- 2019:
-
Saket Singh, Debdoot Sheet, Mithun Dasgupta, "Adversarially Trained Deep Neural Semantic Hashing Scheme for Subjective Search in Fashion Inventory", 2019 4th International Workshop on Fashion and KDD, 25th ACM SIGKDD Conference on Knowledge Discovery and Data Mining
-
Rachana Sathish, Debdoot Sheet, "Unit Impulse Response as an Explainer of Redundancy in a Deep Convolutional Neural Network", Explainable AI Workshop, 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR 2019)
-
Shanka Subhra Mondal, Rachana Sathish, Debdoot Sheet, "Multitask Learning of Temporal Connectionism in Convolutional Networks using a Joint Distribution Loss Function to Simultaneously Identify Tools and Phase in Surgical Videos", MedImage Workshop, 2018 Indian Conference on Vision, Graphics and Image Processing
-
Oindrila Saha, Rachana Sathish, Debdoot Sheet, "Learning with Multitask Adversaries using Weakly Labelled Data for Semantic Segmentation in Retinal Images", International Conference on Medical Imaging with Deep Learning pp. 1-13
-
Ronnie Rajan, Rachana Sathish, Debdoot Sheet, "Significance of Residual Learning and Boundary Weighted Loss in Ischaemic Stroke Lesion Segmentation", 2019 International Conference on Medical Imaging with Deep Learning
-
Oindrila Saha, Rachana Sathish, Debdoot Sheet, "Fully Convolutional Neural Network for Semantic Segmentation of Anatomical Structure and Pathologies in Colour Fundus Images Associated with Diabetic Retinopathy", arXiv preprint arXiv:1902.03122
-
Sumanth Nandamuri, Debarghya China, Pabitra Mitra, Debdoot Sheet, "SUMNet: Fully Convolutional Model for Fast Segmentation of Anatomical Structures in Ultrasound Volumes", 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)
-
Francis Tom, Himanshu Sharma, Dheeraj Mundhra, Tathagato Rai Dastidar, Debdoot Sheet, "Learning a Deep Convolution Network with Turing Test Adversaries for Microscopy Image Super Resolution", 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)
-
Debarghya China, Francis Tom, Sumanth Nandamuri, Aupendu Kar, Mukundhan Srinivasan, Pabitra Mitra, Debdoot Sheet, "UltraCompression: Framework for High Density Compression of Ultrasound Volumes using Physics Modeling Deep Neural Networks", 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019)
-
Kaustuv Mishra, Rachana Sathish, Debdoot Sheet, "Phase Identification and Workflow Modeling in Laparoscopy Surgeries Using Temporal Connectionism of Deep Visual Residual Abstractions", Springer, Cham pp. 201-217
-
Neha Banerjee, Rachana Sathish, Debdoot Sheet, "Deep Neural Architecture for Localization and Tracking of Surgical Tools in Cataract Surgery", Springer, Cham pp. 31-38
-
- 2018:
-
Francis Tom, Debdoot Sheet, "Simulating Patho-realistic Ultrasound Images using Deep Generative Networks with Adversarial Learning", Proc. IEEE Int. Symp. on BiomedicalImaging (ISBI) pp. 1174-1177
-
Debarghya China, Alfredo Illanes, Prabal Poudel, Michael Friebe, Pabitra Mitra, Debdoot Sheet, "Anatomical Structure Segmentation in Ultrasound Volumes using Cross Frame Belief Propagating Iterative Random Walks", IEEE Journal of Biomedical and Health Informatics pp. 1110-1118
-
Aupendu Kar, Sri Phani Krishna Karri, Nirmalya Ghosh, Ramanathan Sethuraman, Debdoot Sheet, "Fully Convolutional Model for Variable Bit Length and Lossy High Density Compression of Mammograms", The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Workshops pp. 2591-2594
-
Kausik Das, Sailesh Conjeti, Abhijit Guha Roy, Jyotirmoy Chatterjee, Debdoot Sheet, "Multiple instance learning of deep convolutional neural networks for breast histopathology whole slide classification", 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) pp. 578-581
-
Prabal Poudel, Alfredo Illanes, Debdoot Sheet, Michael Friebe, "Evaluation of Commonly Used Algorithms for Thyroid Ultrasound Images Segmentation and Improvement Using Machine Learning Approaches", Journal of healthcare engineering pp. 1-13
-
Debarghya China, Pabitra Mitra, Debdoot Sheet, "On the Fly Segmentation of Intravascular Ultrasound Images Powered by Learning of Backscattering Physics", Springer, Cham pp. 351-380
-
Aupendu Kar, Sutanu Bera, SP K Karri, Sudipta Ghosh, Manjunatha Mahadevappa, Debdoot Sheet, "A Deep Convolutional Neural Network Based Classification Of Multi-Class Motor Imagery With Improved Generalization", 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) pp. 5085-5088
-
- 2017:
-
Abhijit Guha Roy, Sailesh Conjeti, Sri Phani Krishna Karri, Debdoot Sheet, Amin Katouzian, Christian Wachinger, Nassir Navab, "ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks", Biomedical optics express pp. 3627-3642
-
Abhijit Guha Roy, Sailesh Conjeti, Debdoot Sheet, Amin Katouzian, Nassir Navab, Christian Wachinger, "Error corrective boosting for learning fully convolutional networks with limited data", International Conference on Medical Image Computing and Computer-Assisted Intervention pp. 231-239
-
Kausik Das, Sri Phani Krishna Karri, Abhijit Guha Roy, Jyotirmoy Chatterjee, Debdoot Sheet, "Classifying histopathology whole-slides using fusion of decisions from deep convolutional network on a collection of random multi-views at multi-magnification", 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017) pp. 1024-1027
-
Hrushikesh Garud, Sri Phani Krishna Karri, Debdoot Sheet, Jyotirmoy Chatterjee, Manjunatha Mahadevappa, Ajoy K Ray, Arindam Ghosh, Ashok K Maity, "High-magnification multi-views based classification of breast fine needle aspiration cytology cell samples using fusion of decisions from deep convolutional networks", Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops pp. 76-81
-
Kaustuv Mishra, Rachana Sathish, Debdoot Sheet, "Learning latent temporal connectionism of deep residual visual abstractions for identifying surgical tools in laparoscopy procedures", Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops pp. 58-65
-
Arna Ghosh, Satyarth Singh, Debdoot Sheet, "Simultaneous localization and classification of acute lymphoblastic leukemic cells in peripheral blood smears using a deep convolutional network with average pooling layer", 2017 IEEE International Conference on Industrial and Information Systems (ICIIS) pp. 1-6
-
S Conjeti, AG Roy, D Sheet, S Carlier, T Syeda-Mahmood, N Navab, A Katouzian, "Domain Adapted Model for In Vivo Intravascular Ultrasound Tissue Characterization", Academic Press pp. 157-181
-
PA Pattanaik, Tripti Swarnkar, Debdoot Sheet, "Object detection technique for malaria parasite in thin blood smear images", 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) pp. 2120-2123
-
- 2016:
-
Debapriya Maji, Anirban Santara, Pabitra Mitra, Debdoot Sheet, "Ensemble of deep convolutional neural networks for learning to detect retinal vessels in fundus images", arXiv preprint arXiv:1603.04833
-
Avisek Lahiri, Abhijit Guha Roy, Debdoot Sheet, Prabir Kumar Biswas, "Deep neural ensemble for retinal vessel segmentation in fundus images towards achieving label-free angiography", 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)
-
Sailesh Conjeti, Amin Katouzian, Abhijit Guha Roy, Loïc Peter, Debdoot Sheet, Stéphane Carlier, Andrew Laine, Nassir Navab, "Supervised domain adaptation of decision forests: Transfer of models trained in vitro for in vivo intravascular ultrasound tissue characterization", Medical image analysis pp. 1-17
-
Abhijit Guha Roy, Sailesh Conjeti, Stephane G Carlier, Pranab K Dutta, Adnan Kastrati, Andrew F Laine, Nassir Navab, Amin Katouzian, Debdoot Sheet, "Lumen segmentation in intravascular optical coherence tomography using backscattering tracked and initialized random walks", IEEE journal of biomedical and health informatics pp. 393-403
-
Pranav Kumar, SL Happy, Swarnadip Chatterjee, Debdoot Sheet, Aurobinda Routray, "An unsupervised approach for overlapping cervical cell cytoplasm segmentation", 2016 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES) pp. 106-109
-
Kausik Basak, Goutam Dey, Manjunatha Mahadevappa, Mahitosh Mandal, Debdoot Sheet, Pranab Kumar Dutta, "Learning of speckle statistics for in vivo and noninvasive characterization of cutaneous wound regions using laser speckle contrast imaging", Microvascular research pp. 6-16
-
Satarupa Banerjee, Swarnadip Chatterjee, Anji Anura, Jitamanyu Chakrabarty, Mousumi Pal, Bhaskar Ghosh, Ranjan Rashmi Paul, Debdoot Sheet, Jyotirmoy Chatterjee, "Global spectral and local molecular connects for optical coherence tomography features to classify oral lesions towards unravelling quantitative imaging biomarkers", RSC Advances pp. 7511-7520
-
Praveen Kumar Kanithi, Jyotirmoy Chatterjee, Debdoot Sheet, "Immersive augmented reality system for assisting needle positioning during ultrasound guided intervention", Proceedings of the Tenth Indian Conference on Computer Vision, Graphics and Image Processing
-
Abhijit Guha Roy, Sailesh Conjeti, Stephane G Carlier, Khalil Houissa, Andreas König, Pranab K Dutta, Andrew F Laine, Nassir Navab, Amin Katouzian, Debdoot Sheet, "Multiscale distribution preserving autoencoders for plaque detection in intravascular optical coherence tomography", 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) pp. 1359-1362
-
Debarghya China, Pabitra Mitra, Debdoot Sheet, "Segmentation of Lumen and External Elastic Laminae in Intravascular Ultrasound Images Using Ultrasonic Backscattering Physics Initialized Multiscale Random Walks", International Conference on Computer Vision, Graphics, and Image processing pp. 393-403
-
Biswajoy Ghosh, Sri Phani Krishna Karri, Debdoot Sheet, Hrushikesh Garud, Arindam Ghosh, Ajoy K Ray, Jyotirmoy Chatterjee, "A generalized framework for stain separation in digital pathology applications", 2016 IEEE Annual India Conference (INDICON) pp. 1-4
-
Manoj Kumar Sharma, Debdoot Sheet, Prabir Kumar Biswas, "Abnormality Detecting Deep Belief Network", Proceedings of the International Conference on Advances in Information Communication Technology & Computing pp. 11
-
Hrushikesh Garud, Sri Phani Krishna Karri, Debdoot Sheet, Ashok Kumar Maity, Jyotirmoy Chatterjee, Manjunatha Mahadevappa, Ajoy Kumar Ray, "Methods and System for Segmentation of Isolated Nuclei in Microscopic Breast Fine Needle Aspiration Cytology Images", International Conference on Computer Vision, Graphics, and Image processing pp. 380-392
-
Kaustuv Mishra, Rachana Sathish, Debdoot Sheet, "Tracking of Retinal Microsurgery Tools Using Late Fusion of Responses from Convolutional Neural Network over Pyramidally Decomposed Frames", International Conference on Computer Vision, Graphics, and Image processing pp. 358-366
-
Manoj Kumar Sharma, Sayan Sarcar, Debdoot Sheet, Prabir Kumar Biswas, "Limitations with measuring performance of techniques for abnormality localization in surveillance video and how to overcome them?", Proceedings of the Tenth Indian Conference on Computer Vision, Graphics and Image Processing pp. 75
-
Suhas Gondi, Kavita Patel, Leslie Mertz, Mary Bates, Jim Banks, Debdoot Sheet, Ahmed Morsy, Jennifer Berglund, Cristian A Linte, Ziv R Yaniv, Michele Solis, "IEEEPULSe", IEEE pp. 1-1
-
Kausik Das, Abhijit Guha Roy, Jyotirmoy Chatterjee, Debdoot Sheet, "Landscaping of random forests through controlled deforestation", 2016 Twenty Second National Conference on Communication (NCC) pp. 1-5
-
- 2015:
-
Debapriya Maji, Anirban Santara, Sambuddha Ghosh, Debdoot Sheet, Pabitra Mitra, "Deep neural network and random forest hybrid architecture for learning to detect retinal vessels in fundus images", 2015 37th annual international conference of the IEEE Engineering in Medicine and Biology Society (EMBC) pp. 3029-3032
-
Debdoot Sheet, Sri Phani Krishna Karri, Amin Katouzian, Nassir Navab, Ajoy K Ray, Jyotirmoy Chatterjee, "Deep learning of tissue specific speckle representations in optical coherence tomography and deeper exploration for in situ histology", 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) pp. 777-780
-
Abhijit Guha Roy, Debdoot Sheet, "Dasa: Domain adaptation in stacked autoencoders using systematic dropout", 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR) pp. 735-739
-
Amrita Chaudhary, Swarnendu Bag, Mousumi Mandal, Sri Phani Krishna Karri, Ananya Barui, Monika Rajput, Provas Banerjee, Debdoot Sheet, Jyotirmoy Chatterjee, "Modulating prime molecular expressions and in vitro wound healing rate in keratinocyte (HaCaT) population under characteristic honey dilutions", Journal of ethnopharmacology pp. 211-219
-
SL Happy, Swarnadip Chatterjee, Debdoot Sheet, "Unsupervised segmentation of overlapping cervical cell cytoplasm", arXiv preprint arXiv:1505.05601
-
Abhijit Guha Roy, Sailesh Conjeti, Stephane G Carlier, Andreas König, Adnan Kastrati, Pranab K Dutta, Andrew F Laine, Nassir Navab, Debdoot Sheet, Amin Katouzian, "Bag of forests for modelling of tissue energy interaction in optical coherence tomography for atherosclerotic plaque susceptibility assessment", 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) pp. 428-431
-
Hrushikesh Tukaram Garud, Debdoot Sheet, Ajoy Kumar Ray, Manjunatha Mahadevappa, Jyotirmoy Chatterjee, "Adaptive weighted-local-difference order statistics filters", US Patent 9208545
-
Sailesh Conjeti, Mehmet Yigitsoy, Debdoot Sheet, Jyotirmoy Chatterjee, Nassir Navab, Amin Katouzian, "Mutually coherent structural representation for image registration through joint manifold embedding and alignment", 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) pp. 601-604
-
Kausik Basak, Goutam Dey, Debdoot Sheet, Manjunatha Mahadevappa, Mahitosh Mandal, Pranab K Dutta, "Probabilistic graphical modeling of speckle statistics in laser speckle contrast imaging for noninvasive and label-free retinal angiography", 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) pp. 6244-6247
-
Sailesh Conjeti, Mehmet Yigitsoy, Tingying Peng, Debdoot Sheet, Jyotirmoy Chatterjee, Christine Bayer, Nassir Navab, Amin Katouzian, "Deformable registration of immunofluorescence and histology using iterative cross-modal propagation", 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI) pp. 310-313
-
Satarupa Banerjee, Debdoot Sheet, Amita Giri, Ranjan Ghosh, Ranjan Rashmi Paul, Mousumi Pal, Jyotirmoy Chatterjee, "optical Coherence Tomographic Attenuation Imaging Based Oral Precancer Diagnosis - Op239.", Head & Neck pp. E133
-
Hrushikesh Tukaram Garud, Debdoot Sheet, Ajoy Kumar Ray, Manjunatha Mahadevappa, Jyotirmoy Chatterjee, "Adaptive Weighted-Local-Difference Order Statistics Filters", US Patent 9208545
-
Shanbao Tong, Debdoot Sheet, Shoaib Bhuiyan, Martha Lucía Zequer Diaz, Andrew Taberne, "BME Trends Around the World: From Baby X to frugal technologies, here's what biomedical engineers are excited about in 2015.[From the Editors]", IEEE pulse pp. 4-6
-
- 2014:
-
Debdoot Sheet, Athanasios Karamalis, Abouzar Eslami, Peter Noël, Jyotirmoy Chatterjee, Ajoy K Ray, Andrew F Laine, Stephane G Carlier, Nassir Navab, Amin Katouzian, "Joint learning of ultrasonic backscattering statistical physics and signal confidence primal for characterizing atherosclerotic plaques using intravascular ultrasound", Medical image analysis pp.103-117
-
Debdoot Sheet, Athanasios Karamalis, Abouzar Eslami, Peter Noël, Renu Virmani, Masataka Nakano, Jyotirmoy Chatterjee, Ajoy K Ray, Andrew F Laine, Stephane G Carlier, Nassir Navab, Amin Katouzian, "Hunting for necrosis in the shadows of intravascular ultrasound", Computerized Medical Imaging and Graphics pp. 104-112
-
Debdoot Sheet, Satarupa Banerjee, Sri Phani Krishna Karri, Swarnendu Bag, Anji Anura, Amita Giri, Ranjan Rashmi Paul, Mousumi Pal, Badal C Sarkar, Ranjan Ghosh, Amin Katouzian, Nassir Navab, Ajoy K Ray, "Transfer learning of tissue photon interaction in optical coherence tomography towardsin vivo histology of the oral mucosa", 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI) pp. 1389-1392
-
Sri Phani Krishna Karri, Hrushikesh Garud, Debdoot Sheet, Jyotirmoy Chatterjee, Debjani Chakraborty, Ajoy Kumar Ray, Manjunatha Mahadevappa, "Learning scale-space representation of nucleus for accurate localization and segmentation of epithelial squamous nuclei in cervical smears", IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI) pp. 772-775
-
Hrushikesh Garud, Debdoot Sheet, Amit Suveer, Manjunatha Mahadevappa, Ajoy Kumar Ray, "Method and apparatus for enhancing representations of micro-calcifications in a digital mammogram image", US Patent 8634630
-
Hrushikesh Garud, Debdoot Sheet, Amit Suveer, Manjunatha Mahadevappa, Ajoy Kumar Ray, "Method and apparatus for enhancing representations of micro-calcifications in a digital mammogram image", US Patent
-